publications
publications by categories in reversed chronological order. generated by jekyll-scholar.
2025
-
 An innovation in host responses to escalating genomic conflictsEmiliano Martí and Amanda M LarracuenteTrends in Genetics, 2025
An innovation in host responses to escalating genomic conflictsEmiliano Martí and Amanda M LarracuenteTrends in Genetics, 2025Conflicts between selfish elements and their hosts can trigger rapid structural and regulatory changes in genomes. Chen et al. discovered a novel species-specific innovation in response to a meiotic driver in Drosophila melanogaster. Their discovery highlights a new dimension in adaptive responses to selfish elements, with broad evolutionary consequences.
@article{marti2025innovation, title = {An innovation in host responses to escalating genomic conflicts}, author = {Mart{\'i}, Emiliano and Larracuente, Amanda M}, journal = {Trends in Genetics}, year = {2025}, publisher = {Cell Press}, doi = {10.1016/j.tig.2025.03.001}, url = {https://authors.elsevier.com/c/1ktQOcQbJF8ya}, dimensions = {true}, } -
 Exploring the Satellitome of the Pest Aphid Acyrthosiphon pisum (Hemiptera, Aphididae): Insights Into Genome Organization and Intraspecies EvolutionLucas Albuquerque, Diogo Milani, Emiliano Martí, and 8 more authorsGenome Biology and Evolution, 2025
Exploring the Satellitome of the Pest Aphid Acyrthosiphon pisum (Hemiptera, Aphididae): Insights Into Genome Organization and Intraspecies EvolutionLucas Albuquerque, Diogo Milani, Emiliano Martí, and 8 more authorsGenome Biology and Evolution, 2025Satellite DNAs (satDNAs), ubiquitous sequences in eukaryotic genomes, play a crucial role in genome organization, function, and evolution. The pea aphid Acyrthosiphon pisum is a major crop pest, and an emerging model for ecological, developmental, and evolutionary studies. This study characterizes the satellitome of the A. pisum to better understand its genomic organization and the evolutionary dynamics of satDNAs at the intraspecies level. The analysis of satDNAs across various host-plant adapted biotypes reveals a general sharing of the satellitome, with significative quantitative differences. We observe the amplification or contraction of specific satDNA families, even in the same biotype, consistent with the fast-evolutionary changes characteristic of these sequences. Rapid differential accumulation of repetitive elements among biotypes together with other factors may contribute to the origin of genetic incompatibilities. At the interspecific level, fluctuations in satDNA abundances, including the loss of certain families, are consistent with the library hypothesis, long-term conservation, and differential amplification of an ancestral satellitome. SatDNAs are considerably enriched in the heterochromatin of the chromosome X (Chr-X) and to a less extent, dispersed across the euchromatic region of all chromosomes. The enrichment of these sequences on Chr-X also features the elements that are most recently amplified and homogenized. This suggests that Chr-X could serve as a fountain of youth for satDNAs, contributing to the “fast-X” effect. Our findings provide valuable insights into genome structure, assembly, and the potential impact of satDNAs on species diversification, as well as the evolution of autosomes and Chr-X.
@article{albuquerque2025apisum, title = {Exploring the Satellitome of the Pest Aphid Acyrthosiphon pisum (Hemiptera, Aphididae): Insights Into Genome Organization and Intraspecies Evolution}, author = {Albuquerque, Lucas and Milani, Diogo and Mart{\'i}, Emiliano and Ferretti, Ana BSM and Rico-Porras, Jos{\'e} Manuel and Mora, Pablo and Pedro, Lorite and Ziabari, Omid S and Brisson, Jennifer A and Palacios-Gimenez, Octavio M and Cabral-de-Mello, Diogo C}, journal = {Genome Biology and Evolution}, year = {2025}, publisher = {Oxford University Press}, doi = {10.1093/gbe/evaf104}, url = {https://academic.oup.com/gbe/article/17/7/evaf104/8196009}, dimensions = {true}, } -
 Genomic origins and evolution of neo-sex chromosomes in Pacific Island birdsChristina A Muirhead, Emiliano Martí, Elsie H Shogren, and 2 more authorsPNAS, 2025
Genomic origins and evolution of neo-sex chromosomes in Pacific Island birdsChristina A Muirhead, Emiliano Martí, Elsie H Shogren, and 2 more authorsPNAS, 2025The standard model of sex chromosome evolution is based on ancient XY or ZW systems. Young neo-sex chromosomes that form via the translocation of autosomal material to preexisting sex chromosomes provide materials to study early events in sex chromosome evolution. Neo-sex chromosomes are taxonomically widespread but, until recently, appeared to be rare in female-heterogametic taxa, including birds. Here, we report on the comparative evolutionary genomics of newly discovered neo-Z and neo-W sex chromosomes in two Pacific Island birds, the honeyeater species, Myzomela tristrami and M. cardinalis. Our results show that 19 to 21 Mya, 85% of chr5, an autosome, translocated to the pseudoautosomal region (PAR) of an ancestral sex chromosome (Z or W) and rapidly recombined onto its heterolog, leaving the remainder of chr5 to segregate as a remnant autosome. A large inversion then established a nonrecombining (NR) neo-W region and a neo-PAR. The accumulation of at least seven additional inversions created interspersed blocks of ancestral and neo-W sequence, without expanding the NR region. The neo-W experienced mass invasion by endogenous retrovirus repeats, a reduced efficacy of selection, and functional degeneration. Many of our observations are consistent with predictions of the standard model, whereas others highlight idiosyncrasies of neo-sex chromosomes that arise via translocation of autosomal sequence to ancestral sex chromosomes. We speculate, in particular, that the repository of retrotransposons on the ancestral W may have been an accelerant in the evolution of neo-W structure, content, and functional degeneration.
@article{muirhead2025myzoneosex, title = {Genomic origins and evolution of neo-sex chromosomes in Pacific Island birds}, author = {Muirhead, Christina A and Mart{\'i}, Emiliano and Shogren, Elsie H and Uy, J Albert C and Presgraves, Daven C}, journal = {PNAS}, volume = {122}, number = {31}, pages = {e2503746122}, year = {2025}, publisher = {National Academy of Sciences}, doi = {10.1073/pnas.2503746122}, url = {https://www.pnas.org/doi/10.1073/pnas.2503746122}, dimensions = {true}, }
2024
-
 Recent secondary contact, genome-wide admixture, and asymmetric introgression of neo-sex chromosomes between two Pacific island bird speciesElsie H Shogren, Jason M Sardell, Christina A Muirhead, and 5 more authorsPLoS genetics, 2024
Recent secondary contact, genome-wide admixture, and asymmetric introgression of neo-sex chromosomes between two Pacific island bird speciesElsie H Shogren, Jason M Sardell, Christina A Muirhead, and 5 more authorsPLoS genetics, 2024Secondary contact between closely related taxa represents a “moment of truth” for speciation—an opportunity to test the efficacy of reproductive isolation that evolved in allopatry and to identify the genetic, behavioral, and/or ecological barriers that separate species in sympatry. Sex chromosomes are known to rapidly accumulate differences between species, an effect that may be exacerbated for neo-sex chromosomes that are transitioning from autosomal to sex-specific inheritance. Here we report that, in the Solomon Islands, two closely related bird species in the honeyeater family—Myzomela cardinalis and Myzomela tristrami—carry neo-sex chromosomes and have come into recent secondary contact after 1.1 my of geographic isolation. Hybrids of the two species were first observed in sympatry 100 years ago. To determine the genetic consequences of hybridization, we use population genomic analyses of individuals sampled in allopatry and in sympatry to characterize gene flow in the contact zone. Using genome-wide estimates of diversity, differentiation, and divergence, we find that the degree and direction of introgression varies dramatically across the genome. For sympatric birds, autosomal introgression is bidirectional, with phenotypic hybrids and phenotypic parentals of both species showing admixed ancestry. In other regions of the genome, however, the story is different. While introgression on the Z/neo-Z-linked sequence is limited, introgression of W/neo-W regions and mitochondrial sequence (mtDNA) is highly asymmetric, moving only from the invading M. cardinalis to the resident M. tristrami. The recent hybridization between these species has thus enabled gene flow in some genomic regions but the interaction of admixture, asymmetric mate choice, and/or natural selection has led to the variation in the amount and direction of gene flow at sex-linked regions of the genome.
@article{shogren2024recent, title = {Recent secondary contact, genome-wide admixture, and asymmetric introgression of neo-sex chromosomes between two Pacific island bird species}, author = {Shogren, Elsie H and Sardell, Jason M and Muirhead, Christina A and Mart{\'i}, Emiliano and Cooper, Elizabeth A and Moyle, Robert G and Presgraves, Daven C and Uy, J Albert C}, journal = {PLoS genetics}, volume = {20}, number = {8}, pages = {e1011360}, year = {2024}, publisher = {Public Library of Science San Francisco, CA USA}, doi = {10.1371/journal.pgen.1011360}, url = {https://journals.plos.org/plosgenetics/article?id=10.1371/journal.pgen.1011360}, dimensions = {true}, }
2023
-
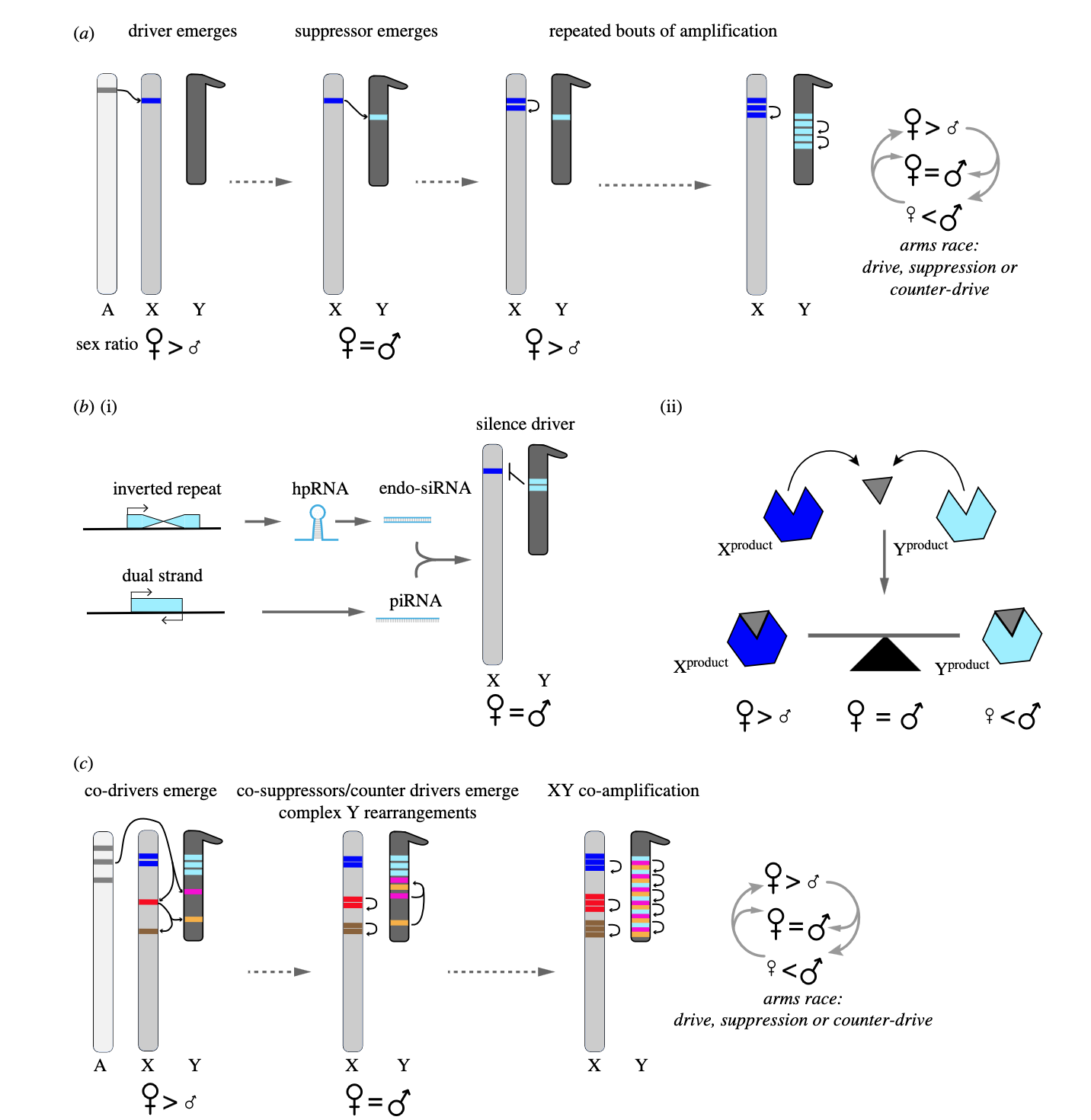 Genetic conflict and the origin of multigene families: implications for sex chromosome evolutionEmiliano Martí and Amanda M LarracuenteProceedings of the Royal Society B, 2023
Genetic conflict and the origin of multigene families: implications for sex chromosome evolutionEmiliano Martí and Amanda M LarracuenteProceedings of the Royal Society B, 2023Sex chromosomes are havens for intragenomic conflicts. The absence of recombination between sex chromosomes creates the opportunity for the evolution of segregation distorters: selfish genetic elements that hijack different aspects of an individual’s reproduction to increase their own transmission. Biased (non-Mendelian) segregation, however, often occurs at a detriment to their host’s fitness, and therefore can trigger evolutionary arms races that can have major consequences for genome structure and regulation, gametogenesis, reproductive strategies and even speciation. Here, we review an emerging feature from comparative genomic and sex chromosome evolution studies suggesting that meiotic drive is pervasive: the recurrent evolution of paralogous sex-linked gene families. Sex chromosomes of several species independently acquire and co-amplify rapidly evolving gene families with spermatogenesis-related functions, consistent with a history of intragenomic conflict over transmission. We discuss Y chromosome features that might contribute to the tempo and mode of evolution of X/Y co-amplified gene families, as well as their implications for the evolution of complexity in the genome. Finally, we propose a framework that explores the conditions that might allow for recurrent bouts of fixation of drivers and suppressors, in a dosage-sensitive fashion, and therefore the co-amplification of multigene families on sex chromosomes.
@article{marti2023genetic, title = {Genetic conflict and the origin of multigene families: implications for sex chromosome evolution}, author = {Mart{\'i}, Emiliano and Larracuente, Amanda M}, journal = {Proceedings of the Royal Society B}, volume = {290}, number = {2010}, pages = {20231823}, year = {2023}, publisher = {The Royal Society}, doi = {10.1098/rspb.2023.1823}, url = {https://royalsocietypublishing.org/doi/full/10.1098/rspb.2023.1823}, dimensions = {true}, }
2022
-
 A step forward in the genome characterization of the sugarcane borer, Diatraea saccharalis: karyotype analysis, sex chromosome system and repetitive DNAs through a cytogenomic approachAna E Gasparotto, Diogo Milani, Emiliano Martí, and 6 more authorsChromosoma, 2022
A step forward in the genome characterization of the sugarcane borer, Diatraea saccharalis: karyotype analysis, sex chromosome system and repetitive DNAs through a cytogenomic approachAna E Gasparotto, Diogo Milani, Emiliano Martí, and 6 more authorsChromosoma, 2022Moths of the family Crambidae include a number of pests that cause economic losses to agricultural crops. Despite their economic importance, little is known about their genome architecture and chromosome evolution. Here, we characterized the chromosomes and repetitive DNA of the sugarcane borer Diatraea saccharalis using a combination of low-pass genome sequencing, bioinformatics, and cytogenetic methods, focusing on the sex chromosomes. Diploid chromosome numbers differed between the sexes, i.e., 2n = 33 in females and 2n = 34 in males. This difference was caused by the occurrence of a WZ1Z2 trivalent in female meiosis, indicating a multiple sex-chromosome system WZ1Z2/Z1Z1Z2Z2. A strong interstitial telomeric signal was observed on the W chromosome, indicating a fusion of the ancestral W chromosome with an autosome. Among repetitive DNAs, transposable elements (TEs) accounted for 39.18% (males) to 41.35% (females), while satDNAs accounted for only 0.214% (males) and 0.215% (females) of the genome. FISH mapping revealed different chromosomal organization of satDNAs, such as single localized clusters, spread repeats, and non-clustered repeats. Two TEs mapped by FISH were scattered. Although we found a slight enrichment of some satDNAs in the female genome, they were not differentially enriched on the W chromosome. However, we found enriched FISH signals for TEs on the W chromosome, suggesting their involvement in W chromosome degeneration and differentiation. These data shed light on karyotype and repetitive DNA dynamics due to multiple chromosome fusions in D. saccharalis, contribute to the understanding of genome structure in Lepidoptera and are important for future genomic studies.
@article{gasparotto2022step, title = {A step forward in the genome characterization of the sugarcane borer, Diatraea saccharalis: karyotype analysis, sex chromosome system and repetitive DNAs through a cytogenomic approach}, author = {Gasparotto, Ana E and Milani, Diogo and Mart{\'i}, Emiliano and Ferretti, Ana Beatriz SM and Bardella, Vanessa B and Hickmann, Frederico and Zrzav{\'a}, Magda and Marec, Franti{\v{s}}ek and Cabral-de-Mello, Diogo C}, journal = {Chromosoma}, volume = {131}, number = {4}, pages = {253--267}, year = {2022}, publisher = {Springer Berlin Heidelberg Berlin/Heidelberg}, doi = {10.1007/s00412-022-00781-4}, url = {https://link.springer.com/article/10.1007/s00412-022-00781-4}, dimensions = {true}, }
2021
-
 New insights into the six decades of Mesa’s hypothesis of chromosomal evolution in Ommexechinae grasshoppers (Orthoptera: Acridoidea)Mylena D Santander, Diogo C Cabral-de-Mello, Alberto Taffarel, and 4 more authorsZoological Journal of the Linnean Society, 2021
New insights into the six decades of Mesa’s hypothesis of chromosomal evolution in Ommexechinae grasshoppers (Orthoptera: Acridoidea)Mylena D Santander, Diogo C Cabral-de-Mello, Alberto Taffarel, and 4 more authorsZoological Journal of the Linnean Society, 2021In Acridoidea grasshoppers, chromosomal rearrangements are frequently found as deviations from the standard acrocentric karyotype (2n = 23♂/24♀, FN = 23♂/24♀) in either phylogenetically unrelated species or shared by closely related ones, i.e. genus. In the South American subfamily Ommexechinae, most of the species show a unique karyotype (2n = 23♂/24♀, FN = 25♂/26♀) owing to the occurrence of a large autosomal pair (L1) with submetacentric morphology. In the early 1960s, Alejo Mesa proposed the hypothesis of an ancestral pericentric inversion to explain this karyotype variation. Furthermore, in Ommexechinae, extra chromosomal rearrangements (e.g. centric fusions) are recorded between the ancestral X chromosome and autosomes that originated the so-called neo-sex chromosomes. However, the evolutionary significance of the pericentric inversions and centric fusions in Ommexechinae remains poorly explored. Aiming for a better understanding of chromosomal evolution in Ommexechinae, we performed a detailed cytogenetic analysis in five species. Our findings support the hypothesis about the occurrence of an early pericentric inversion in the ancestor of Ommexechinae. Moreover, our results show a complex karyotype diversification pattern due to several chromosome rearrangements, variations in heterochromatin and repetitive DNA dynamics. Finally, the chromosomal mapping of U2 snDNA in L1 provided new insights about the morphological evolution of this autosomal pair and revealed unnoticed chromosome reorganizations.
@article{santander2021new, title = {New insights into the six decades of Mesa's hypothesis of chromosomal evolution in Ommexechinae grasshoppers (Orthoptera: Acridoidea)}, author = {Santander, Mylena D and Cabral-de-Mello, Diogo C and Taffarel, Alberto and Mart{\'i}, Emiliano and Mart{\'i}, Dardo A and Palacios-Gimenez, Octavio M and Castillo, Elio Rodrigo D}, journal = {Zoological Journal of the Linnean Society}, volume = {193}, number = {4}, pages = {1141--1155}, year = {2021}, publisher = {Oxford University Press UK}, doi = {10.1093/zoolinnean/zlaa188}, url = {https://academic.oup.com/zoolinnean/article/193/4/1141/6138200}, dimensions = {true}, } -
 Cytogenomic analysis unveils mixed molecular evolution and recurrent chromosome rearrangements shaping the multigene families on Schistocerca grasshopper genomesEmiliano Martí, Diogo Milani, Vanessa B Bardella, and 4 more authorsEvolution; international journal of organic evolution, 2021
Cytogenomic analysis unveils mixed molecular evolution and recurrent chromosome rearrangements shaping the multigene families on Schistocerca grasshopper genomesEmiliano Martí, Diogo Milani, Vanessa B Bardella, and 4 more authorsEvolution; international journal of organic evolution, 2021Multigene families are essential components of eukaryotic genomes and play key roles either structurally and functionally. Their modes of evolution remain elusive even in the era of genomics, because multiple multigene family sequences coexist in genomes, particularly in large repetitive genomes. Here, we investigate how the multigene families 18S rDNA, U2 snDNA, and H3 histone evolved in 10 species of Schistocerca grasshoppers with very large and repeat-enriched genomes. Using sequenced genomes and fluorescence in situ hybridization mapping, we find substantial differences between species, including the number of chromosomal clusters, changes in sequence abundance and nucleotide composition, pseudogenization, and association with transposable elements (TEs). The intragenomic analysis of Schistocerca gregaria using long-read sequencing and genome assembly unveils conservation for H3 histone and recurrent pseudogenization for 18S rDNA and U2 snDNA, likely promoted by association with TEs and sequence truncation. Remarkably, TEs were frequently associated with truncated copies, were also among the most abundant in the genome, and revealed signatures of recent activity. Our findings suggest a combined effect of concerted and birth-and-death models driving the evolution of multigene families in Schistocerca over the last 8 million years, and the occurrence of intra- and interchromosomal rearrangements shaping their chromosomal distribution. Despite the conserved karyotype in Schistocerca, our analysis highlights the extensive reorganization of repetitive DNAs in Schistocerca, contributing to the advance of comparative genomics for this important grasshopper genus.
@article{marti2021cytogenomic, title = {Cytogenomic analysis unveils mixed molecular evolution and recurrent chromosome rearrangements shaping the multigene families on Schistocerca grasshopper genomes}, author = {Mart{\'i}, Emiliano and Milani, Diogo and Bardella, Vanessa B and Albuquerque, Lucas and Song, Hojun and Palacios-Gimenez, Octavio M and Cabral-de-Mello, Diogo C}, journal = {Evolution; international journal of organic evolution}, volume = {75}, number = {8}, pages = {2027--2041}, year = {2021}, publisher = {Oxford University Press}, doi = {10.1111/evo.14287}, url = {https://academic.oup.com/evolut/article/75/8/2027/6730336}, dimensions = {true}, } -
 Genomic differences between the sexes in a fish species seen through satellite DNAsCarolina Crepaldi, Emiliano Martí, Évelin Mariani Gonçalves, and 2 more authorsFrontiers in Genetics. More Information can be found here , 2021
Genomic differences between the sexes in a fish species seen through satellite DNAsCarolina Crepaldi, Emiliano Martí, Évelin Mariani Gonçalves, and 2 more authorsFrontiers in Genetics. More Information can be found here , 2021Neotropical fishes have highly diversified karyotypic and genomic characteristics and present many diverse sex chromosome systems, with various degrees of sex chromosome differentiation. Knowledge on their sex-specific composition and evolution, however, is still limited. Satellite DNAs (satDNAs) are tandemly repeated sequences with pervasive genomic distribution and distinctive evolutionary pathways, and investigating satDNA content might shed light into how genome architecture is organized in fishes and in their sex chromosomes. The present study investigated the satellitome of Megaleporinus elongatus, a freshwater fish with a proposed Z1Z1Z2Z2/Z1W1Z2W2 multiple sex chromosome system that encompasses a highly heterochromatic and differentiated W1 chromosome. The species satellitome comprises of 140 different satDNA families, including previously isolated sequences and new families found in this study. This diversity is remarkable considering the relatively low proportion that satDNAs generally account for the M. elongatus genome (around only 5%). Differences between the sexes in regards of satDNA content were also evidenced, as these sequences are 14% more abundant in the female genome. The occurrence of sex-biased signatures of satDNA evolution in the species is tightly linked to satellite enrichment associated with W1 in females. Although both sexes share practically all satDNAs, the overall massive amplification of only a few of them accompanied the W1 differentiation. We also investigated the expansion and diversification of the two most abundant satDNAs of M. elongatus, MelSat01-36 and MelSat02-26, both highly amplified sequences in W1 and, in MelSat02-26’s case, also harbored by Z2 and W2 chromosomes. We compared their occurrences in M. elongatus and the sister species M. macrocephalus (with a standard ZW sex chromosome system) and concluded that both satDNAs have led to the formation of highly amplified arrays in both species; however, they formed species-specific organization on female-restricted sex chromosomes. Our results show how satDNA composition is highly diversified in M. elongatus, in which their accumulation is significantly contributing to W1 differentiation and not satDNA diversity per se. Also, the evolutionary behavior of these repeats may be associated with genome plasticity and satDNA variability between the sexes and between closely related species, influencing how seemingly homeologous heteromorphic sex chromosomes undergo independent satDNA evolution.
@article{crepaldi2021genomic, title = {Genomic differences between the sexes in a fish species seen through satellite DNAs}, author = {Crepaldi, Carolina and Mart{\'i}, Emiliano and Gon{\c{c}}alves, {\'E}velin Mariani and Mart{\'i}, Dardo Andrea and Parise-Maltempi, Patricia Pasquali}, journal = {Frontiers in Genetics}, volume = {12}, pages = {728670}, year = {2021}, publisher = {Frontiers Media SA}, doi = {10.3389/fgene.2021.728670}, url = {https://www.frontiersin.org/journals/genetics/articles/10.3389/fgene.2021.728670}, dimensions = {true}, }
